A1: An introduction to what one can learn from biomimetics as exemplified by PQQ-Ln complexes
The still recent discovery of the biological relevance of lanthanides (Ln) has opened the door to numerous new ideas and topics that utilize nature’s methods for chemical transformations. Since the characterization of the exclusively Ln-dependent methanol dehydrogenase (MDH) in Methylacidiphilum fumariolicum (SolV) by Pol et al. in 2014,[1] studies on biomimetic complexes of the enzyme have been aiming towards a better understanding of the mechanism in the biological system and towards the development of efficient catalysts for the oxidation of alcohols.
Although Ln3+ ions have been extensively used as probes for Ca2+ in biochemistry (similar ionic radii and coordination numbers, but favourable spectroscopic properties), they were not thought to have any biological relevance for a long time.[2] In 2011, Kawai et al. discovered that the methylotroph1 Methylorubrum extroquens (AM1) contained not only a Ca2+ - dependent methanol dehydrogenase, but also a Ln3+ - dependent MDH for metabolizing methanol to formaldehyde.[3] It was shown that even nanomolar concentrations are sufficient to trigger the expression of Ln-MDH. In addition, a bacterium that solely relies on lanthanides was discovered in 2014 by Pol et al.: Methylacidiphilum fumariolicum (SolV).[1] The group around Pol showed that this organism could not grow with Ca2+ alone in the medium and that the growth increased from Gd to Eu to Sm and reached a maximum with the early lanthanides La, Ce, Nd or Pr. Further investigations showed that SolV can only express the Ln-dependent MDH and has no option so switch to a Ca-MDH, explaining why Ln are absolutely essential for the energy metabolism of this organism. The authors also provided a crystal structure of this Ln-MDH which revealed a pyrroloquinoline quinone (PQQ, Figure 1) cofactor and a lanthanide ion in its active site. Compared to the crystal structure of the Ca-MDH,[4] the Ln-MDH additionally contains an aspartate residue in the active site to satisfy the higher coordination number of the lanthanides.
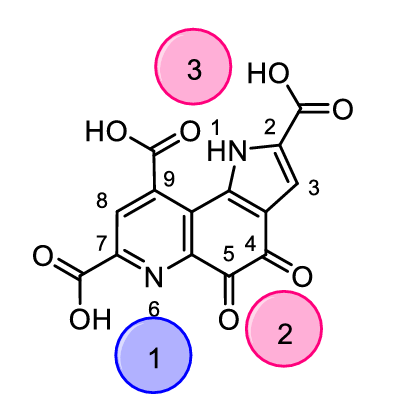
s intriguing as the newly discovered field of lanthanide biocatalysis in MDHs may be, several aspects of the catalytic system remain to be elucidated. Limited research on lanthanide models has been performed until now, only enabling fragmented information of the processes occurring at the reaction site. Several aspects such as the preference for early lanthanides while later ones are less efficient and the exact steps that take place between metal center, cofactor, substrate and solvent molecules are still to be investigated. The answers to these questions will facilitate the development of effective bioinspired lanthanide catalysts.
Hoping to better understand the mechanism of alcohol oxidation in MDH, several research groups have been investigating biomimetic complexes, focusing on the search of useful ligand platforms. Some of these investigated ligands are depicted in Figure 2.
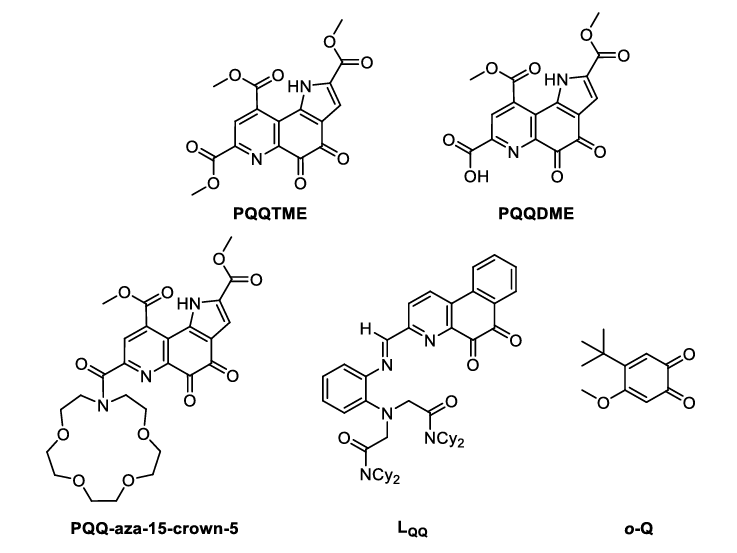
In combination with a suitable metal ion (lanthanide or calcium), these ligands are able to perform the conversion of alcohols to aldehydes in a similar manner to the enzyme they mimic. With different spectroscopic methods, one is able to assess the reactivity of these complexes, which includes the substrate turnover properties and the changes that undergo at the cofactor (ligand) site. In this case, biomimetics serves two purposes:
- Mechanism elucidation: compared to their enzymatic counterparts, spectroscopic analysis of small molecule biomimetic complexes is far easier to prepare and interpret. With a larger amount of available analysis methods, more information can be gained about the molecular processes taking place.
- Biomimetic catalysis: the reaction being catalyzed by an enzyme takes place in its active site. Biomimetic complexes, which closely resemble the molecular structure of the active site, are often able to facilitate the same reaction catalyzed by the enzyme itself. However, with small molecule complexes being synthetically accessible, the effort to achieve the catalyst is far smaller than in the case of the enzyme.
In the following, a short overview will be given over the methods that the authors of the studies use and to what purposes they use them.
UV-Vis spectroscopy: As the cofactor PQQ and the ligands derived from it show specific absorption bands in the UV and visible regions, reactions taking place at the PQQ moiety can be followed by UV-Vis spectroscopy. For example, the complexation of metal ions to the ligand, the addition of nucleophiles to the quinone moiety or the reduction of the quinone during substrate oxidation can be followed using this method.[6c, 6d, 6f]
EPR spectroscopy: With the help of EPR spectroscopy, the authors of the o-Q study have been able to show that the mechanism of substrate oxidation takes place via a radical intermediate detectable by this method.[6f]
NMR spectroscopy: Apart from the usual characterization of ligands and complexes using 1H-NMR spectroscopy, Schelter and coworkers and Daumann and coworkers have used this method to quantify the amount of aldehyde product formed by their catalyst by comparison of the integrals of the product signals with an added internal standard.[6e, 6g] Alternatively, GC-MS has been used by Luo and coworkers for product quantification.[7]
Mass spectrometry: In their 2021 study, Daumann and Weis used mass spectrometry coupled ion mobility to investigate the size/structure of the complexes as well as their stability with different counterions.[6g, 8]
With these tools in hand, one can gather data about the system regarding its structure as well as its reactivity with the aim to better understand the biological system being mimicked and also to hopefully be able to develop catalysts which can be applied to a large scope of substrates.
1 A bacterium that uses C1 carbon compounds in its energy metabolism.
References
[1] A. Pol, T. R. M. Barends, A. Dietl, A. F. Khadem, J. Eygensteyn, M. S. M. Jetten, H. J. M. Op den Camp, Environ. Microbiol. 2014, 16, 255–264.
[2] aJ. Reuben, Sci. Nat. 1975, 62, 172–178; bE. Nieboer, in Rare Earths, Vol. 22, Springer Berlin Heidelberg, Berlin, Heidelberg, 1975, pp. 1–47; cP. Mulqueen, J. M. Tingey, W. D. Horrocks, Biochemistry 1985, 24, 6639–6645.
[3] aY. Hibi, K. Asai, H. Arafuka, M. Hamajima, T. Iwama, K. Kawai, J. Biosci. Bioeng. 2011, 111, 547–549; bN. A. Fitriyanto, M. Fushimi, M. Matsunaga, A. Pertiwiningrum, T. Iwama, K. Kawai, J. Biosci. Bioeng. 2011, 111, 613–617; cT. Nakagawa, R. Mitsui, A. Tani, K. Sasa, S. Tashiro, T. Iwama, T. Hayakawa, K. Kawai, PloS One 2012, 7, e50480.
[4] P. A. Williams, L. Coates, F. Mohammed, R. Gill, P. T. Erskine, A. Coker, S. P. Wood, C. Anthony, J. B. Cooper, Acta Crystallogr. D Biol. Crystallogr. 2005, 61, 75–79.
[5] N. Nakamura, T. Kohzuma, H. Kuma, S. Suzuki, Inorg. Chem. 1994, 33, 1594–1599.
[6] aS. Itoh, M. Mure, Y. Ohshiro, T. Agawa, Tetrahedron Lett. 1985, 26, 4225-4228; bS. Itoh, H. Kawakami, S. Fukuzumi, J. Am. Chem. Soc. 1997, 119, 439–440; cS. Itoh, H. Kawakami, S. Fukuzumi, Biochemistry 1998, 37, 6562–6571; dS. Itoh, H. Kawakami, S. Fukuzumi, J. Mol. Catal., B Enzym. 2000, 8, 85–94; eA. McSkimming, T. Cheisson, P. J. Carroll, E. J. Schelter, J. Am. Chem. Soc. 2018, 140, 1223-1226; fR. Zhang, R. Zhang, R. Jian, L. Zhang, M.-T. Zhang, Y. Xia, S. Luo, Nat. Commun. 2022, 13, 428; gV. A. Vetsova, K. R. Fisher, H. Lumpe, A. Schäfer, E. K. Schneider, P. Weis, L. J. Daumann, Chem. Eur. J. 2021, 27, 10087–10098.
[7] C. Anthony, L. J. Zatman, Biochem. J. 1967, 104, 960–969.
[8] A. Schafer, V. A. Vetsova, E. K. Schneider, M. Kappes, M. Seitz, L. J. Daumann, P. Weis, J. Am. Soc. Mass Spectrom. 2022, 33, 722-730.

